 ^^^
^^^
Group of Membrane Physiology and Bioenergetics

Scientist
PD Dr. Jost Ludwig
Postdoctoral Fellow
MSc. Katsiaryna Shamayeva, PhD.
Graduate Student
MSc. Oksana Munko
Research Assistant
Ing. Karin Spurná
PD Dr. Jost Ludwig
Phone: +420 389 033 844
ludwig@nh.cas.cz
Address
Zamek 136, 37333 Nove Hrady
Research
Structural and functional analysis of the yeast K+ translocation system(s) encoded by TRK1 and TRK2 Genes
Researchers involved from the group: Deepika Kale, Karin Spurna, Pavel Spurny, Kastiarina Shamayeva, Jost LudwigInternal collaboration: Natallia Kulik, Saurabh Kumar Pandey, Rudiger H. Ettrich,
External collaboration: Vasilina Zayats (University of Warsaw, Poland) and Thmas Stockner (Medical University, Vienna, Austria)
Project Aim:
Potassium (K+) ions are the most abundant ion species in living cells. In non-excitable cells, K+ provides favorable conditions for many enzymatic reactions and serves as osmoticum. In excitable cells, action potentials and signal transduction as well as modulation of signals are usually based on electric currents carried by K+ (and Na+ and Ca2+) ions. In mammalian cells, which are in a controlled and rather constant environment, K+ homeostasis is maintained by the Na+/K ATPase. Sessile organisms like yeast (fungi) and plant as well as prokaryotes have to cope with K+ that can change in several orders of magnitude while intracellular K+ has to be kept (more or less) constant. For that, a variety of cation translocation systems in plasma- and organellar membranes developed (for review see e.g. Epstein 2003 (bacteria); Gierth & Mäser 2007; Grabov 2007 (plants); Ariño et al. 2010 (yeast)). Since the K+ translocation across membranes in prokaryotes, plants and fungi strongly differs from that in mammals, its investigation could also be valuable for developing drugs against pathogenic species. The yeast Saccharomyces cerevisiae is not only one of the most important biotechnologically (i.e. in the production of bioethanol) used eukaryotic microorganism, but also serves as model to study (among many other cellular processes) membrane transport. Its worth to note here that recently, it has been demonstrated that an increased activity of K+ translocation systems in turn increases tolerance to ethanol and thus the yield in bioethanol production (Lam et al. 2014).
In yeast TRK1 and TRK2 genes encode K+ uptake system(s) that allow cells to grow in media containing only a few μM K+. Trk1 and Trk2 belong to the SKT family of ion translocation systems that have evolved from a precursor of prokaryotic K-channels. Most SKT proteins have acquired cytoplasmic regulatory domains encoded by a another gene, while Trk1,2 possess a “long hydrophilic loop” (LHL) inserted in the peptide chain within the transmembrane domain. Although, the sequence of Trk1,2 proteins is known since 1991, the structure is not yet available. Except of their main role, K+ transport, Trks are able to translocate Cl-.which most probably requires multimerization of several Trk1,2 proteins. The project is based on our previous work, in which we developed a refined model of the transmembrane part of Trk1. It is designed as a combined experimental and theoretical (molecular modeling) approach to study the multimerization state of Trk1,2 proteins including possible heteromer formation, the role of multimerization in K+ or Cl- translocation and to define the role of LHL. 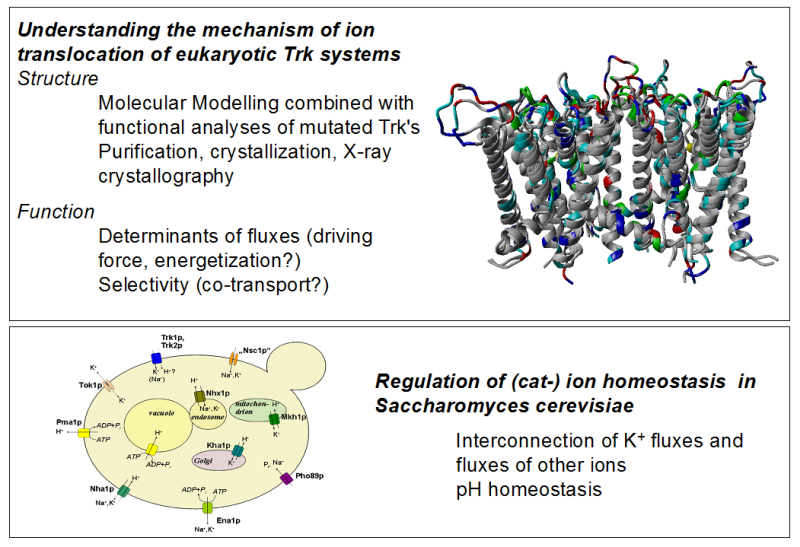
- Selected recent publications from our group
- [1] Vasilina Zayats, Thomas Stockner, Saurabh K Pandey, Katharina Wörz, Rüdiger Ettrich, Jost Ludwig (2015) A refined atomic scale model of the Saccharomyces cerevisiae K+-translocation protein Trk1p combined with experimental evidence confirms the role of selectivity filter glycines and other key residues. BBA-Biomembranes 1848 (5): 1183-1195.
Subprojects:
Structural feature of a yeast potassium translocation protein examined with MD simulations and electrostatics calculations
Researchers involved from the group: Jost LudwigInternal collaboration: Natallia Kulik, Saurabh Kumar Pandey, Rudiger H. Ettrich,
External collaboration: Vasilina Zayats (University of Warsaw, Poland)
Potassium channels are crucial in the life of a yeast cell, allowing them to grow in media with low concentrations of the cation.1 Trk1 is one such transmembrane protein. There is evidence to suggest that chloride transport is also associated with this protein; however, the mechanism is not well understood as the quaternary structure is not fully understood. Through homology modeling, a dimeric structure of Trk1 was proposed. This structure does not include any features that would initially indicate a chloride transport; a dimeric form was proposed because the model was recrystallized as a dimer. Therefore, it is not known whether Trk1 exists in the cell as a monomer, dimer, or tetramer.
Through electrostatic calculations, the four different domains of Trk1 are examined to determine which would be most likely to interact. Several models (monomer, dimers, and tetramers) are proposed and simulated in cell-like conditions for up to 500ns. The stability of each complex is determined, as well as their potassium cation and chloride anion affinities. In addition, the path of potassium translocation is determined to elucidate the transport mechanism for ions.
Biochemical Analysis of Membrane Proteins in Yeast
Researchers involved from the group: Deepika Kale, Jost LudwigProject Aim:
Analysis of the S. cerevisiae TRK proteins using biochemical methods (SDS-PAGE, Western blot).
Short Annotation:
TRK proteins are cation translocating proteins that allow S. cerevisiae (S.c) to survive and grow under different environment from a few µM to hundreds of mM [K+] and maintaining internal K+ concentration relatively constant. Intracellular K+ plays a pivotal role in essential cellular processes, such as osmotic regulation, protein synthesis and enzyme activities. It is also required activation of key metabolic processes and for negative charge compensation. In S.c there are two specific K+ translocation systems: Trk1 and Trk2. Trk1 is 1235 amino acids long and Trk2 is 889 amino acid long. A unique feature of fungal Trks is that they contain a “Long Hydrophilic Loop” (LHL) which is not homologous between these proteins and differs largely in length i.e. 648 aa in Trk1 and 327 aa in Trk2. In this project analysis of full length Trk’s and LHL of Trk1 and Trk2 will be carried out using different biochemical methods. Main aim is the standardisation of a protocol for the isolation of Trk1 and Trk2 from yeast membranes, and to isolate LHL from the cytosol where different protocols to isolate membrane proteins and cytosolic proteins will be performed. Isolated protein concentration will be determined using Bradford’s method. Analysis of isolated protein fractions (Trk1, Trk2, Trk1/LHL and Trk2/LHL) by SDS-PAGE, followed by western blot analysis. Methods used within this project will also include microbiological techniques like maintenance and growth of yeast cells and fluorescence microscopy to examine the cellular localisation of Trk/GFP fusion proteins.
Expression, Purification (and Crystallization) of Protein Fragments
Researchers involved from the group: Katsiarina Shamayeva, Jost LudwigPotassium (K+) ions are the most abundant ion species in almost all living cells. In non-excitable cells, K+ e.g. provides favorable conditions for many enzymatic reactions and serves as osmoticum. Baker’s yeast (Saccharomyces cerevisiae) cells are able to grow in environments with a wide range of external [K+] ranging from less than 100 μM to more than 1 M. Internal [K+] is maintained relatively constant by a variety of cation transport systems in the plasma – and organelle membranes of S. cerevisiae. In S. cerevisiae there are two specific K+translocation systems: Trk1 and Trk2. Both are large membrane proteins made out of 1235 (Trk1) and 889 (Trk2) amino acid residues, respectively. The crystal structure of Trk-proteins is not known. Based on related proteins, we have developed an atomic scale molecular model that describes the core region (e.g. the membrane spanning parts) of Trk. However this model cannot provide information about several cytosolic parts of Trk that show no homology to any other proteins.
Within this project we aim to express and purify different fragments of TRK and set up crystallization experiments using a crystallization robot. The techniques used will be transformation of bacterial (E. coli) cells, growth of cultures and protein purification using FPLC (ÄKTA) for affinity (e.g. His-tag) and size exclusion chromatography.
Bimolecular Fluorescence Complementation as a Tool to Determine Composition of Multimeric Yeast Membrane Proteins
Researchers involved from the group: Karin Spurna, Jost LudwigProject Aim:
Determining of the composition of multimeric yeast membrane proteins by Bimolecular Fluorescence Complementation.
Short Annotation:
TRK1 encodes the main potassium translocation systems in Saccharomyces cerevisiae. TRK encodes on one polypeptide chain four domains that are homologous to (inwardly rectifying) potassium channel subunits that are functional as tetramer. Thus, the protein could be in vivo functional as monomer. However, it seems more likely that Trk is either dimer or tetramer.
To determine possible interactions between proteins and protein subunits, Bimolecular Fluorescence complementation (BiFC) can be used. This technique is based on the ability of some proteins to form functional complexes when their polypeptide chain is split into two parts and if these two parts are in close proximity. In this project split “Venus” (a variant of the green fluorescent protein, GFP) will be used to analyse interactions between Trk1 subunits. The amino-terminal and carboxy-terminal parts of Venus will be fused to Trk1 or partial deletion mutants by molecular biology methods. Yeast strains co-“expressing” these proteins will be generated and analysed by fluorescence microscopy. Fluorescence should only occur if these “half-proteins” are close enough to allow constitution of full Venus.
Non-Invasive Measurements of Ion Fluxes Across Cell Membranes
Researchers involved from the group: Pavel Spurny, Jost LudwigProject Aim:
Using “Microelectrode based flux estimation (MIFE)” and “Flux estimation with ion selective electrodes” (FLISE) for measuring fluxes of specific ions across the plasma membrane of yeast cells.
Short Annotation:
In order to determine ionic currents across cell membranes, the patch clamp technique that allows very precise measurements is widely used. Whereas patch clamp is almost always the method of choice when currents (ion fluxes) across mammalian cells are analysed, it can be problematic with cells that possess cell walls (e.g. plant and fungi cells). In that case removal of this cell wall that prohibits the giga-seal formation is necessary. This works rather well with many cells, but can be problematic e.g. with yeast cells. In any case, the treatment might change the behaviour of ion channels and transporters. Within the project two alternative, non-invasive methods for determination of ion fluxes are used that both rely on measuring changes of ion concentrations in the surrounding of cells:
“Microelectrode based flux estimation (MIFE)” which was developed in the Biophysics laboratory at the University of Tasmania is based on ion-selective microelectrodes which are moving between two positions (closer and further) above cell monolayer or another cell structure. Ion fluxes are calculated from the small differences of concentrations between these two positions. MIFE can be used with organs, tissues (or tissue slices and cell layers). Within the project, MIFE is used to determine ion fluxes using monolayers of yeast cells. 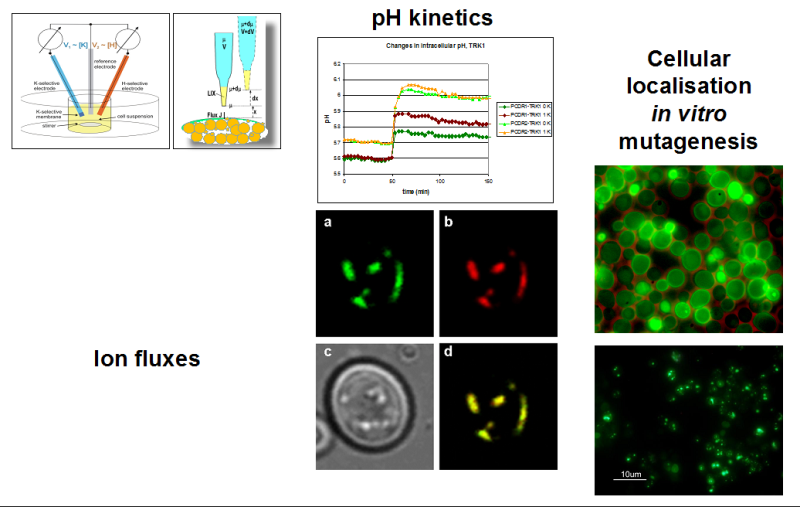 “Flux measurements with Ion Selective Electrodes” (FLISE), developed in our lab, depends on measuring global ion concentration changes. It is used to measure transmembrane fluxes of K+, Na+ and H+ with suspensions of yeast cells expressing (mutants of) TRK1, the main cation translocation system in yeast.
“Flux measurements with Ion Selective Electrodes” (FLISE), developed in our lab, depends on measuring global ion concentration changes. It is used to measure transmembrane fluxes of K+, Na+ and H+ with suspensions of yeast cells expressing (mutants of) TRK1, the main cation translocation system in yeast.



